Capturing the molecular choreography of carbon fixation
Article Highlights
- Plant Research Laboratory researchers disprove a long-held view on the permeability of carboxysome shells using molecular dynamics
- In between the proteins that assemble the carboxysome shell, minute gaps exist that gas can move through
- Understanding these shells may allow researchers to engineer cyanobacteria or plants to photosynthesize more efficiently
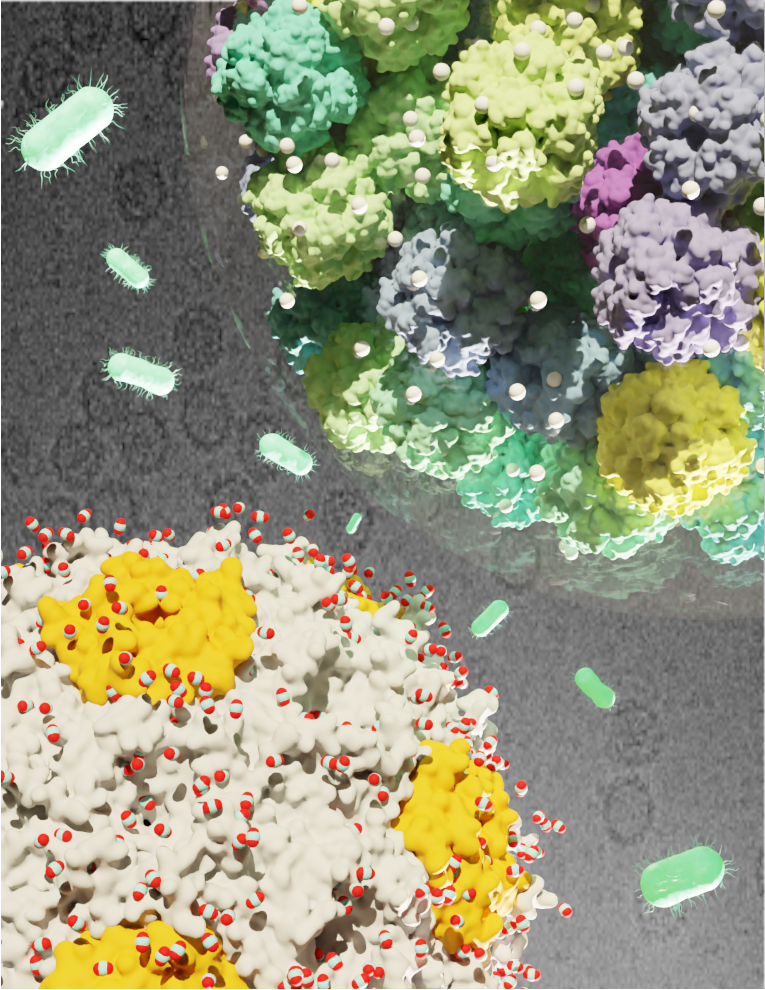
Multiscale modeling of carbon fixation in cyanobacteria, depicted in aquagreen. Michigan
State University researchers are using molecular dynamics to better understand photosynthesis.
Image by: Daipayan Sarkar, Christopher M. Maffeo, Markus Sutter, Aleksei Aksimentiev,
Cheryl A. Kerfeld, Josh V. Vermaas
Using molecular dynamics, researchers disprove a long-held view on the permeability of carboxysome shells, revealing a new path for both carbon dioxide and research in photosynthesis.
Photosynthesis, the process by which plants turn sunlight into food, is vital for all life on Earth. Plants aren’t the only ones who photosynthesize, and cyanobacteria are of particular interest to researchers studying how photosynthesis works.
Within the photosynthetic machinery of cyanobacteria are structures known as carboxysome shells, which capture carbon dioxide from the atmosphere. CO2 is brought into the shell – in the form of bicarbonate – to start the process of building the molecules of life known as carbon fixation.
Carboxysome shells are built like soccer balls, with flat hexagonal and pentagonal shapes made of proteins that assemble into a shell. Initially, it was believed that these shells were gas tight, with any traffic of gas coming in and out controlled by central pores in the shell.
However, research from the Michigan State University-Department of Energy Plant Research Laboratory, or PRL, shows that where the flat shapes touch, minute gaps exist where gases can escape through. This research was published in The Proceedings of the National Academy of Sciences.
“We saw that there are some alternate pathways and energetically confirmed that there are for gas diffusion, especially CO2, which is critical for the function for photosynthesis,” said Daipayan Sarkar, postdoctoral researcher in the Vermaas lab and first author of the study.
The researchers used computer simulations known as molecular dynamics to analyze the transport of gases going in and out of carboxysome shells.
“For me, this paper gave me a visual image of molecules in motion reflecting off the protein shell,” said Cheryl Kerfeld, Hannah Distinguished Professor at the PRL and in the MSU Department of Biochemistry and Molecular Biology, or BMB, and author on this study. “As a structural biologist, my point of view is static images of structures; working with Josh and Daipayan adds dynamics and accordingly, new insight.”
The researchers also ran a second simulation, where they packed a carboxysome shell with rubisco, the enzyme the grabs CO2 for photosynthesis. Simulations of gas diffusion in crowded carboxisomes were performed using software Atomic Resolution Brownian Dynamics, developed by Christopher Maffeo and Aleksei Aksimentiev at University of Illinois Urbana-Champaign.

By Kara Headley
“The dynamic picture is helping us understand how small molecules permeate across these large molecular assemblies,” said Sarkar.
Imagine a room full of dancers, the rubisco takes the lead, and is looking for a partner, or CO2, to dance with. The CO2 can either dance with the rubisco or leave the ballroom, through the gaps in the shell. Interestingly, the researchers found that the CO2 will quickly find a dance partner rather than leave alone.
The shell, then, makes carbon fixation much more efficient, despite offering escape routes for carbon dioxide.
“The shell makes the turnover-to-leave ratio about 100x better than without a shell present, so you can really beautifully see how evolution would select for this otherwise awkward-looking shell system,” said Josh Vermaas, assistant professor in the PRL and BMB, and principal investigator on this study.
Going forward, the researchers still have plenty of mysteries to uncover when it comes to the carboxysome shell.
“What is very important right now, especially for the PRL, is meeting the DOE’s sustainability goals by addressing fundamental questions in basic energy sciences,” Sarkar said. “In this work we revisited the fundamental question on metabolite permeability, and using molecular simulations we show how synthetic microcompartments can be used for carbon fixation applications.”
Plants and photosynthetic bacteria, known as cyanobacteria, can be genetically engineered to have more of these shells, which would in turn make them more efficient at photosynthesis.
“Based on the modularity of these microcompartments, you can now make or design technologies where you can express them with an enzyme of your choice, and then utilize it for carbon fixation or other sustainability challenges we collectively face as a society,” Sarkar said.
The work in this article is supported by the US Department of Energy, Office of Basic Energy Sciences under grant number DE-FG02-91ER20021.



