NEEDLE points to gene regulators: Q&A with Federica Brandizzi
User-friendly platform predicts which proteins trigger gene expression, enabling scientists to engineer more productive and hardy crops
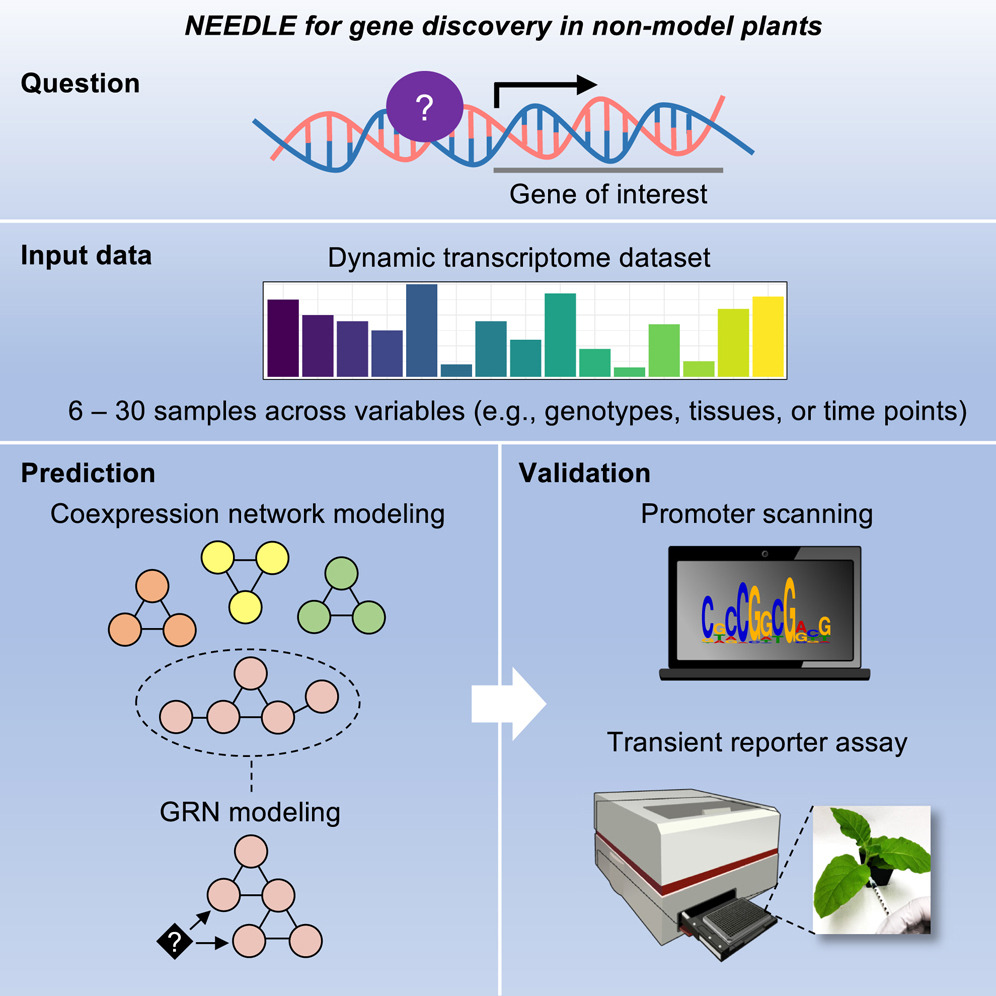
Transcription factors are proteins that bind to DNA inside a cell, activating or blocking the expression of a particular gene. Accurately predicting these gene regulators is a key step to making more productive and stress-resistant crops.
But the complex interactions between DNA, RNA and proteins within cells make this difficult, and scientists lack comprehensive datasets for most crop species. Therefore, scientists with the MSU-DOE Plant Research Laboratory, or PRL, developed a user-friendly pipeline, known colloquially as NEEDLE, to identify the transcription factors that regulate target genes associated with important traits.
The following is a Q&A with Federica Brandizzi, the lead PI on this project, NEEDLE.

Credit: MSU
How might NEEDLE be applicable to the PRL or the Department of Energy Basic Energy Sciences (BES) research?
NEEDLE could be highly relevant to PRL and BES research in multiple ways. The ability to construct gene regulatory networks (GRNs) from transcriptomic data aligns well with PRL’s emphasis on understanding photosynthetic efficiency, stress responses and metabolic pathways in photosynthetic organisms. For BES, NEEDLE’s application in identifying transcription factors (TFs) that regulate critical biosynthetic pathways could accelerate the discovery of genes involved in photosynthesis in crops. For example, the approach used to uncover CSLF6 regulators in grasses could be adapted to study genes involved in carbon assimilation, energy capture, conversion, and storage at all scales, or resilience to environmental stress - key topics in PRL and BES-funded research. NEEDLE could also facilitate the integration of multi-omics data for non-model species, supporting PRL’s long-term goal of discovering how photosynthetic organisms work on the molecular level.
What, to you, is the most exciting part of this research?
The most exciting aspect of NEEDLE is its ability to systematically identify transcriptional regulators in non-model species using only transcriptomic data, eliminating the need for extensive multi-omics datasets. The integration of co-expression network analysis with GRN prediction and in planta validation provides a powerful, streamlined approach to gene discovery. The study’s application of NEEDLE to CSLF6 regulation in Brachypodium and sorghum is particularly compelling, because it not only identifies novel regulators, but also reveals evolutionary divergence in transcriptional control mechanisms. This comparative approach could be extended to a wide range of plant traits, offering insights into how regulatory networks evolve across species.
Is there anything else you would like to add?
NEEDLE’s minimal computational requirements and user-friendly workflow make it a valuable tool for plant scientists without extensive bioinformatics expertise. This idea emerged from the need to make analyses of large datasets on crops easily accessible and workable for researchers. I am particularly proud of Dr. Dae Kwan Ko, who designed the pipeline and successfully implemented it, as demonstrated in this work. While the study demonstrates its effectiveness, the ability to integrate additional datasets (e.g., epigenomic data, metabolic profiles) could further enhance its predictive power. Additionally, applying NEEDLE to stress-related regulatory networks in bioenergy crops or photosynthetic model organisms at the PRL could provide valuable insights into optimizing plant productivity under changing environmental conditions. If coupled with CRISPR-based validation strategies, NEEDLE could help accelerate functional characterization and translational applications of key regulatory genes in crop improvement.
To learn more about NEEDLE, visit the Great Lakes Bioenergy Research Center’s newsroom. Sections of this press release were adapted from a GLBRC research highlight by Chris Hubbuch.
This study was supported primarily by the Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research with contributing support from National Institutes of Health Chemical Sciences, Geoscience and Biosciences Division; DOE Office of Basic Energy Sciences; and MSU AgBioResearch.



