Considering multiple binding sites for proteins with domain similar to the small subunit of rubisco
Protein modeling in cyanobacteria predicts binding interactions between rubisco and proteins with homology to the small subunit of rubisco.
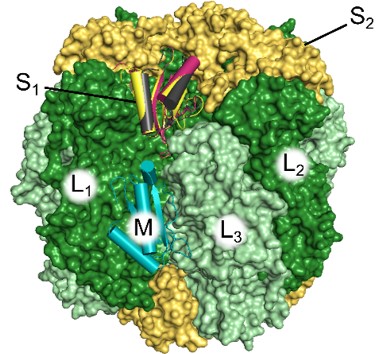
Scientific Achievement
Protein homology modeling in cyanobacteria predicts binding interactions, at two sites, between rubisco and proteins with a domain (SSLD) similar to the small subunit of rubisco.
Significance and Impact
Rubisco fixes carbon dioxide (CO2) during the process of photosynthesis. The study suggests that SSLDs could bind at additional rubisco sites than previously shown. This variety suggests dynamic regulatory mechanisms to be explored regarding these protein-protein interactions.
Research Details
- The small subunit-like domain is found in two proteins in cyanobacteria and is important for their interaction with rubisco.
- They may bind in the same location as the rubisco small subunit, since they are homologous to the small subunit of rubisco, though recent studies suggest that the binding is at a different site.
- Modeling of the full small subunit-like domains in an additional organism supports a view that considers binding at both sites.
- The small subunit-like domain is not expected to bind the S1 position as well as RbcS, but it still forms favorable interactions at this site and maintains many important interactions, especially when considering residues that were absent in published experimental studies.
- Previous work studied environments that would favor the new M binding site, but other conditions, species, and post-translational modifications could allow for binding at the small subunit site as well.
Related people: Brandon Rohnke, Cheryl Kerfeld, Beronda Montgomery (CA)
This work was primarily funded by the US Department of Energy, Office of Basic Energy Sciences.



