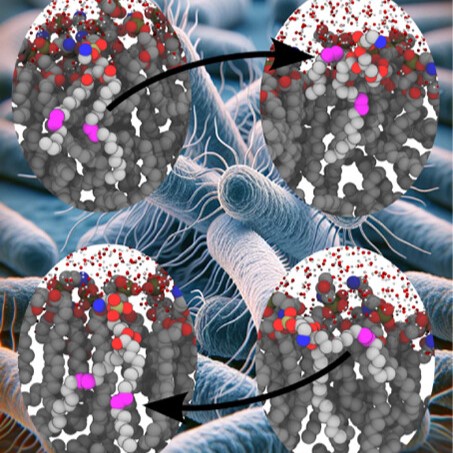
Exploring cryo-electron microscopy with molecular dynamics
Integrating single particle cryo-electron microscopy (EM) and molecular dynamics (MD) has made a paradigm shift to the field of structural biology in order to determine both structure and function of biomolecules.
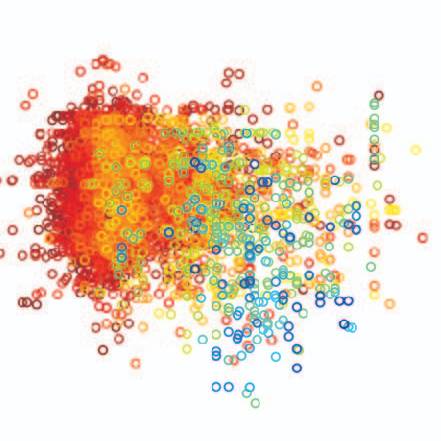
Recent increases in market share among PDB structures for cryo-EM methods highlighted in (A) are driven by the increasing accessibility for high-resolution electron densities for large biological macromolecules (B). The underlying data for panels (A) and (B) are taken from the Protein Data Bank, accessed October 2021.
By John Vant
Scientific Achievement
In this review article, we present a current overview of integrative modeling in a post resolution revolution era in cryo-EM. With the advancement in detector technology, a wide range of computational methods – both physics based and using machine learning – have been developed to determine high resolution structures of proteins in diverse biological systems such as plants, bacteria and viruses. Some of these computational methods include molecular dynamics flexible fitting (MDFF), cross-correlation driven molecular dynamics (CDMD), Cryo-BIFE, MultiMap and many more as listed under Table 1 of the review.
Significance and Impact
In addition to the methods, the review tabulates (Table 2) some of the recent exemplary examples where integrative modeling methods have been adopted to determine a high-resolution map and model of biological systems critical to human health. Such recent examples include the high-resolution structure of the Chimpanzee Adenovirus (ChAdOx1) Oxford-AstraZeneca COVID-19 vaccine at 3.07 Å and the spike protein structure of the coronavirus (SARS-COV-2) at 3.46 Å, to name a few.
Research Details
Our collaborators from Abhishek Singharoy's group at Arizona State University and Josh Vermaas’s lab at the MSU-DOE Plant Research Laboratory were invited by Biochemical Society Transactions to work on a special issue of the journal focusing on computational biology, molecular dynamics and structural biology. Although we recognize that there are different experimental and computational methods in the field of structural biology, in this review we focus on the success of integrating cryo-EM and molecular dynamics, as we anticipate that this will be the future of experimental and computational structural biology.
Related people: John W. Vant, Daipayan Sarkar, Jonathan Nguyen, Alexander T. Baker, Josh V. Vermaas and Abhishek Singharoy
DOI: 10.1042/BST20210485
This work was funded in part by the US Department of Energy, Office of Basic Energy Sciences.