Scientists discover potential mechanism of ultra-rare blood clots linked to adenovirus COVID-19 vaccines
An international team of scientists, including Michigan State University researchers, believe they may have found a molecular mechanism behind the extremely rare blood clots linked to adenovirus COVID-19 vaccines.
The global team used state-of-the-art technology to analyze the AstraZeneca vaccine in minute detail to understand whether the ultra-rare side effect could be linked to the viral vector.

Image Credit: Daipayan Sarkar/Josh Vermaas
Their findings, which were recently published in the international journal Science Advances, suggest it is the viral vector – in this case an adenovirus used to shuttle the coronavirus’ genetic material into cells – and the way it binds to platelet factor 4 (PF4) once injected that could be the potential mechanism that triggers blood clots in a very small number of people after the vaccine is administered.
Researchers from Cardiff University and Arizona State University worked with AstraZeneca scientists to investigate vaccine-induced immune thrombotic thrombocytopenia (VITT), also known as thrombosis with thrombocytopenia syndrome (TTS), a life-threatening condition seen in a very small number of people after receiving the Oxford-AstraZeneca or Johnson & Johnson vaccines.
“VITT only happens in extremely rare cases because a chain of complex events needs to take place to trigger this ultra-rare side effect,” said Alan Parker, an expert in the use of adenoviruses for medical applications from Cardiff University’s School of Medicine and first author of the study. “Our data confirms PF4 can bind to adenoviruses, an important step in unravelling the mechanism underlying VITT.”
When both vaccines showed the ultra-rare side effect of VITT, scientists wondered whether the viral vector had some part to play. Another important clue was that neither the Moderna nor Pfizer vaccines, made from an entirely different technology called mRNA vaccines, showed this effect.

Courtesy photo
MSU’s role in the research was in refining the cryo-electron microscopy (cryo-EM) structures, integrating the collective wisdom from four decades of biomolecular simulation into the structure generation pipeline. Using the initial cryo-EM structures as a template, the MSU team conducted molecular simulations for large fractions of the viral capsid proteins using the electron densities from cryo-EM as an additional bias to our simulations.
“The key advance we made was to incorporate the inherent capsid symmetry into the modeling process, preventing unphysical geometries that might otherwise occur once a full viral capsid is built,” said Josh Vermaas, a computational biophysicist in the MSU Department of Biochemistry and Molecular Biology in the College of Natural Science, an MSU-DOE Plant Research Laboratory faculty member and study co-author. “Once elements corresponding to poorly resolved density were removed by our collaborators, the resulting structures were uploaded to the protein data bank for other researchers to use in their own structural studies for these vaccine vectors.”

Courtesy photo
Daipayan Sarkar, a postdoctoral research associate in the Vermaas lab and study co-author, acknowledged the contributions of all members who participated in the international transatlantic research effort, especially in the middle of the pandemic.
“I also greatly appreciated the unique opportunity and the innumerable scientific/technical discussions we had as we pushed toward higher resolution,” Sarkar added.
In particular, the team detailed the structure and receptor of ChAdOx1, which is adapted from chimpanzee adenovirus Y25 – and how it interacts with PF4. They believe it is this specific interaction – and how it is then presented to the immune system – that could prompt the body’s own defenses to view it as foreign and release of antibodies against this self-protein.
The research team used computational models, which the Singharoy group at ASU specialize in, to show that one of the ways the two molecules tightly bind is via electrostatic interactions.
Credit: Daipayan Sarkar/Josh Vermaas
“We found that ChAdOx1 has a strong negative charge,” Baker said. “This means the viral vector can act like a magnet and attract proteins with the opposite, positive charge, like PF4. We then found that PF4 is just the right size and shape that when it gets close to ChAdOx1 it could bind in between the negatively charged parts of ChAdOx1’s surface, called hexons.”
The research team is hopeful that, armed with a better understanding of what may be causing rare VITT, they can provide further insights into how vaccines and other therapies, which rely on the same technology, might be altered in the development of the next generation vaccines and therapies.
“Although very rare, it is critical we fully investigate vector-host interactions of the vaccine at a mechanistic level to help us understand both how the vaccine generates immunity and how it may lead to any rare adverse events, such as VITT,” Baker said. “Establishing a mechanism could help to prevent and treat this disorder.
"This project reflects some of the best trends in science since the pandemic began, with really wide-ranging collaborations across institutions, bringing together expertise that goes beyond an individual laboratory or institution,” Vermaas added. “I’m really excited to see how we can use these techniques on other symmetric viruses and organelles across biology, and couple more closely experiment and simulation."
The Medicines and Healthcare products Regulatory Agency (MHRA) continues to advise that vaccination is the best way to protect people from COVID-19 and that the benefits far outweigh the risk of any known side effects.
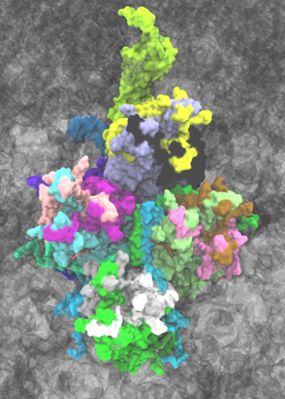
Banner image: Atomic model developed for the Chimpanzee Adenovirus viral vector used in the AstraZeneca COVID-19 vaccine. The virus is made up of repeating proteins related by symmetry, with each of the 60 symmetry units shown as a different color (point cloud in the background). The individual major (hexon, penton) and minor capsid proteins (pIIIa, pVI, pVIII, pIX) within an asymmetric unit are shown with different colors within the central unit in surface representation. The figure on the right zooms into a single asymmetric unit fitted in a cryo-EM density, represented as grey cloud around the individual atoms within the structure. Image was rendered in VMD. Credit: Daipayan Sarkar/Josh Vermaas
Read the original story in the College of Natural Science newsroom.



